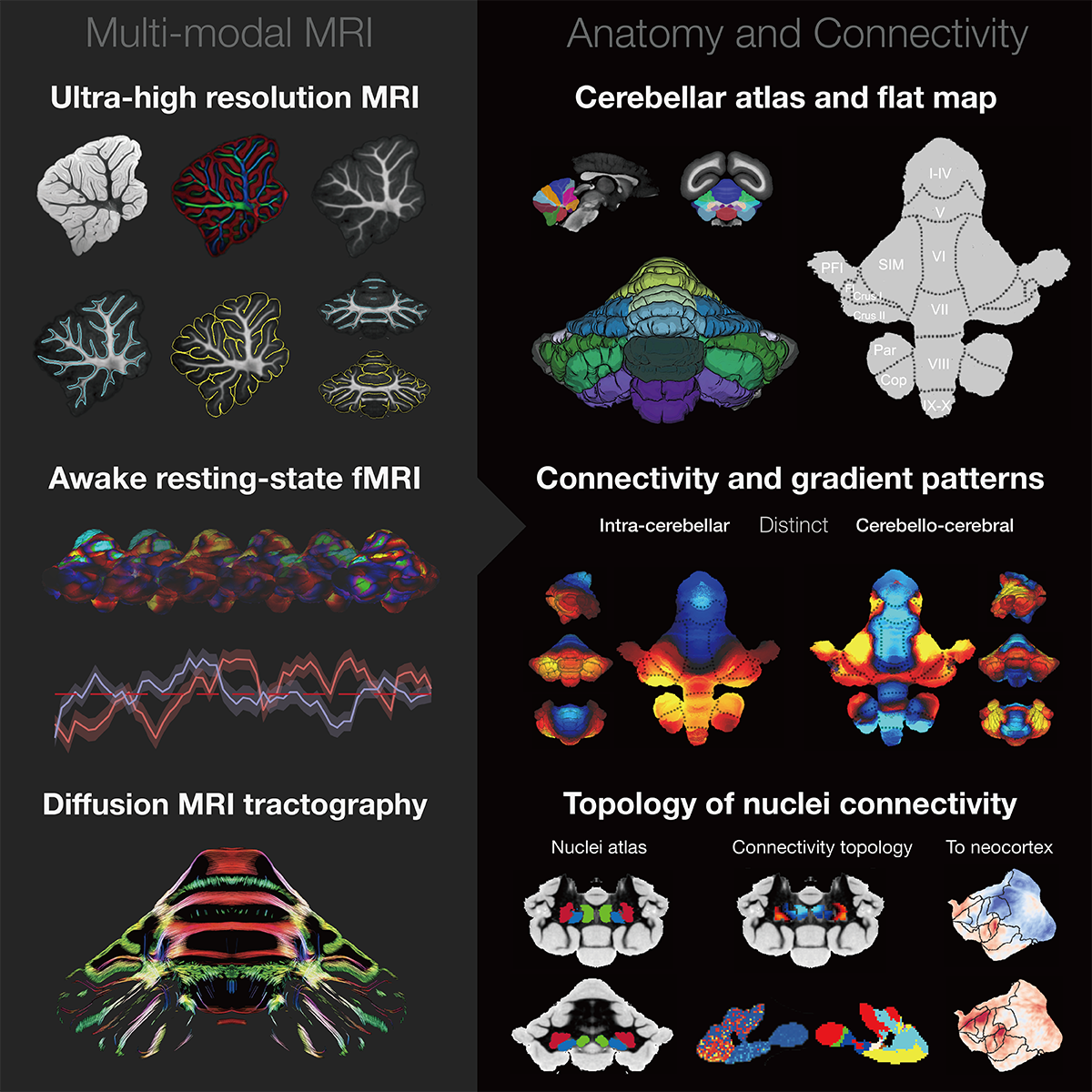
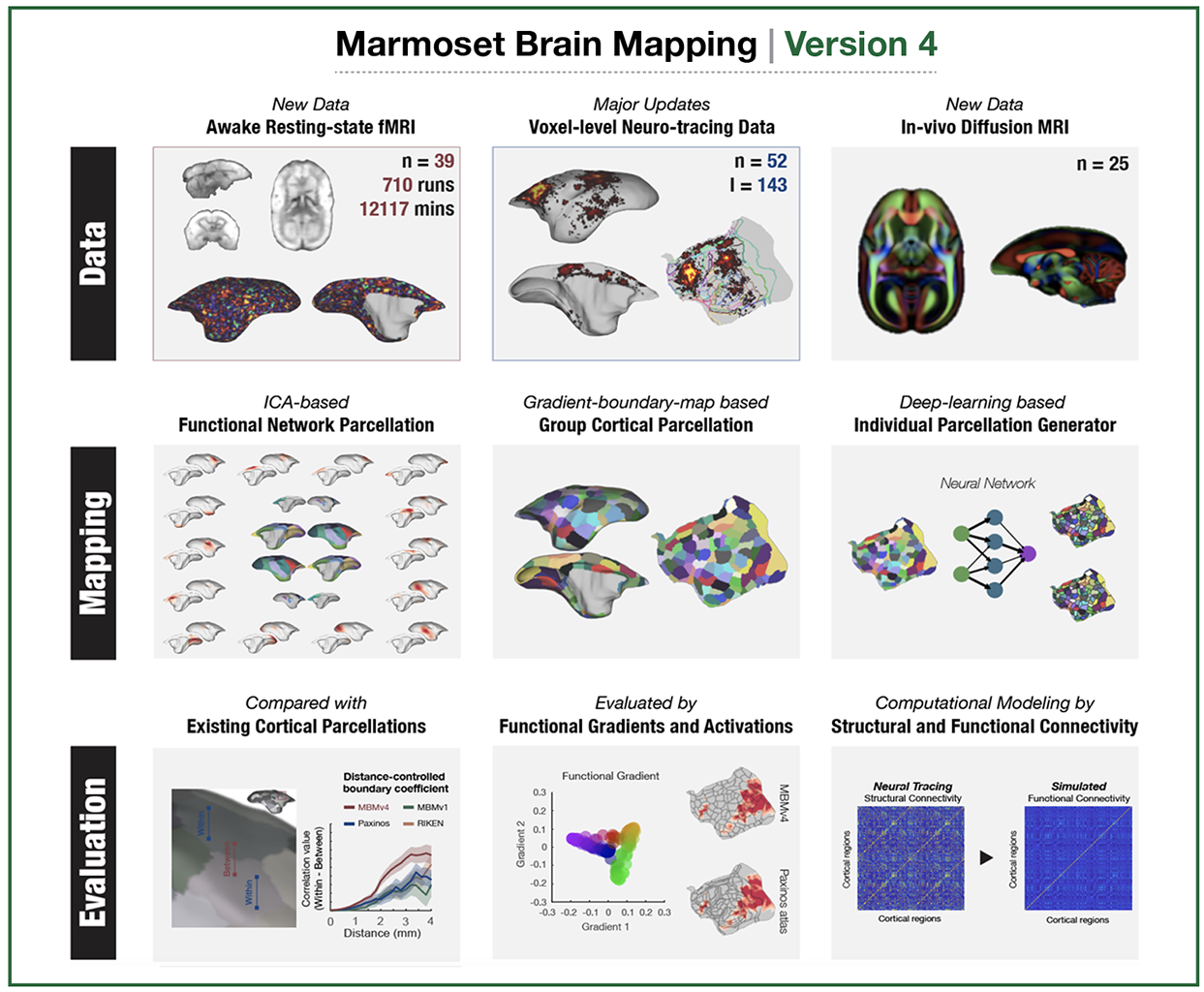
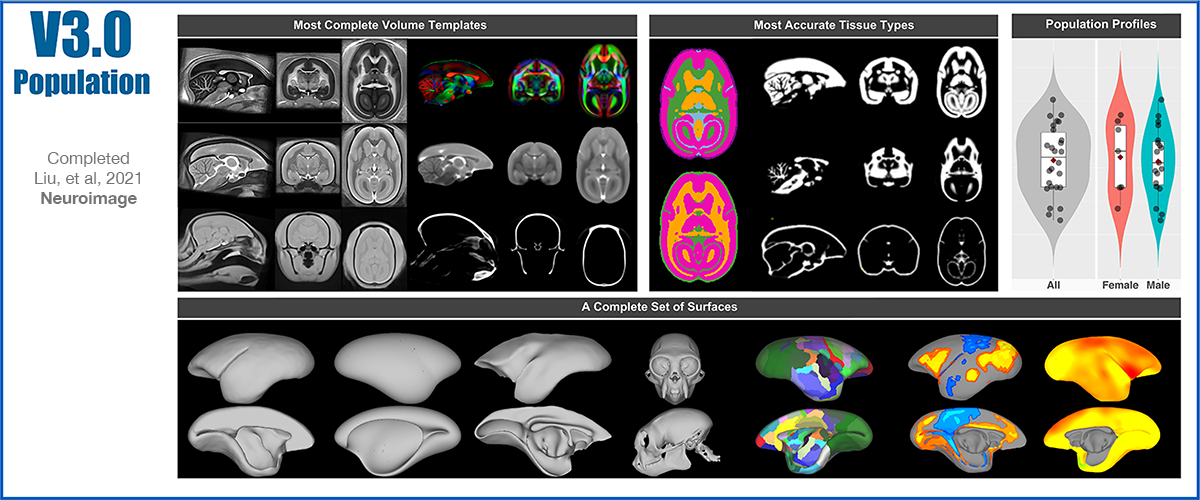
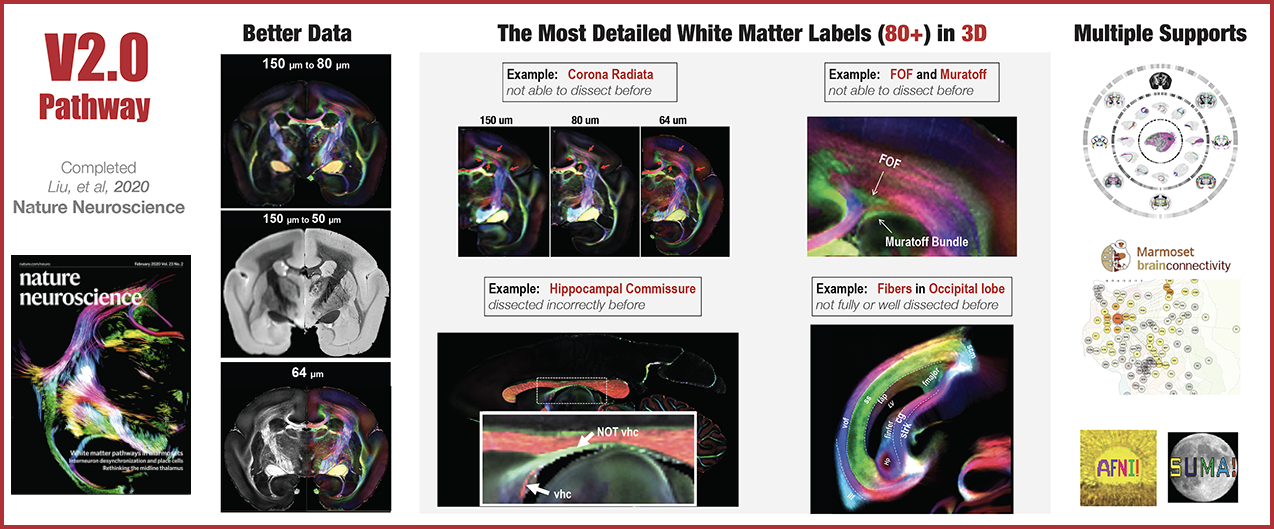
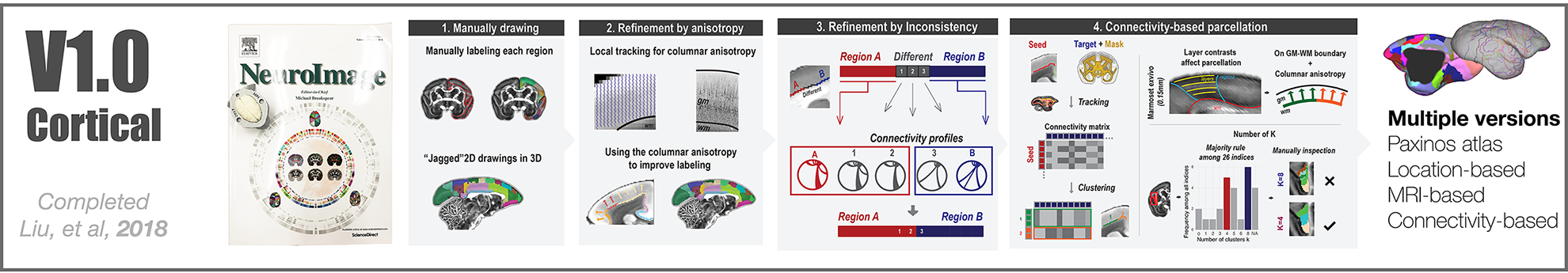
The Version-5 part-2 Focused on the Cerebellum Spatial Transcriptome
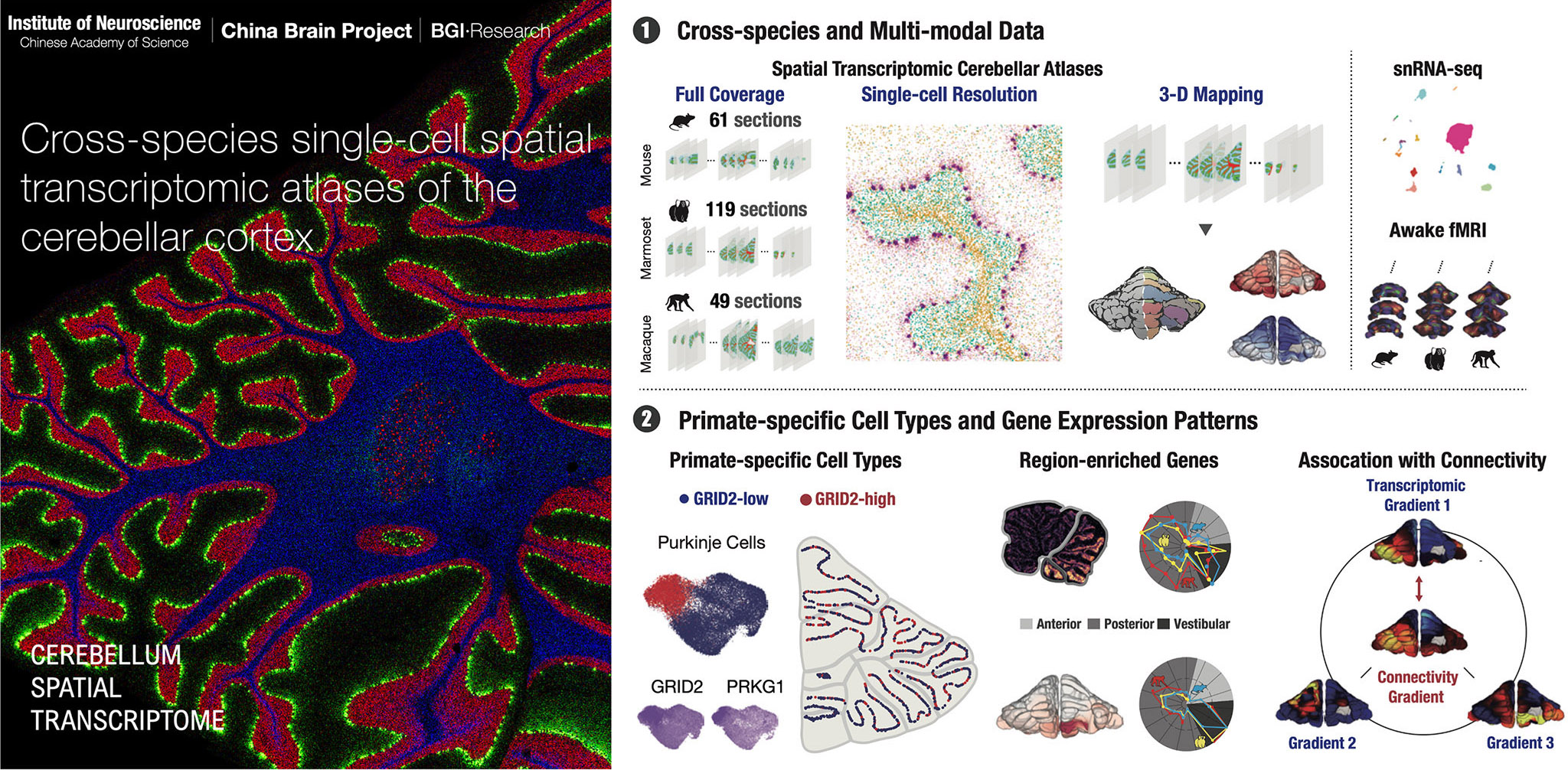
Hao, et al. An Anatomical and Connectivity Atlas of the Marmoset Cerebellum. Science, 2024
Abstract The molecular and cellular organization of the primate cerebellum remains poorly characterized. We obtained single-cell spatial transcriptomic atlases of macaque, marmoset, and mouse cerebella and identified primate-specific cell subtypes, including Purkinje cells and molecular-layer interneurons, that show different expression of the glutamate ionotropic receptor Delta type subunit 2 (GRID2) gene. Distinct gene expression profiles were found in anterior, posterior, and vestibular regions in all species, whereas region-selective gene expression was predominantly observed in the granular layer of primates and in the Purkinje layer of mice. Gene expression gradients in the cerebellar cortex matched well with functional connectivity gradients revealed with awake functional magnetic resonance imaging, with more lobule-specific differences between primates and mice than between two primate species. These comprehensive atlases and comparative analyses provide the basis for understanding cerebellar evolution and function.