CIRONG LIU LAB @ ION
Marmoset Brain Mapping | Neuroanatomy | Connectome | Transcriptome
Cirong LIU, Ph.D. Investigator
Research Interests Lab of Translational Brain Imaging Institute of Neuroscience Chinese Academy of Sciences crliu@ion.ac.cn or cirongliu@gmail.com
Xin Wang Lab Manager
Surgery | Behavior 一针见血
Yafeng Zhan Postdoc
Connectome | Neuroimaging
Zefang Wang Research Assistant
那本珍贵的笔记本
Jiankang Xi PhD Student
Electrophysiology | Behavior自带BKB
Qihang Wu PhD Student
Computation | Connectome燃起来!
Yao Fei Visiting Student
Connectome | Neuroimaging
Tao Song PhD Student
Connectome | Transcriptome 我导愿称我为最强!
Kemeng Xu Master Student
Bioinformatics
Ao Fu Visiting Student
Behavior | Electrophysiology 手戳电极 无人能敌
Jixuan Lin Master Student
Bioinformatics | Transcriptome yysy“🤪”真不戳!!
Haichao Qu Visiting Student
Multi-modal Integration | NeuroAI
Mingkai Yao Master Student
Marmoset Behavior
Ximeng Liu Master Student
Neuroimaging
Xiuying Zhang PhD Student
Neuroimaging
Yu Zhang Visiting Student
NeuroAI
Simiao Zhang Visiting Student
Connectome
Postdoc and RA
Lijing Zhu/RA Hean Liu/RA Haotian Yang/RA
Graduate Students
Song Kun/PhD
Visiting Students
Wenyuan Li Chuanyao Wei Xiaojia Zhu Boyang Zhang Qiyu Wang Kun Jiang Haonan Zhao
Undergraduate and Interns
Xiaorui Liu Furui Feng Boshuang Wang Hongjuan Hu
Fei T#, Dong Y#, Gou X#, Zhang J#, Wang G#, Yuan N#, Zhuang Z#, Chen D#, ..., Liu S*, Liu C*, Shen Z*, Sun Y*, Li C*, Mulder J*, Cao G*, Sun Y*, Xu C* Evolutionary convergence and divergence of hippocampal cytoarchitecture between rodents and primates revealed by single-cell spatial transcriptomics (2026) National Science Review (*Co-corresponding authors)
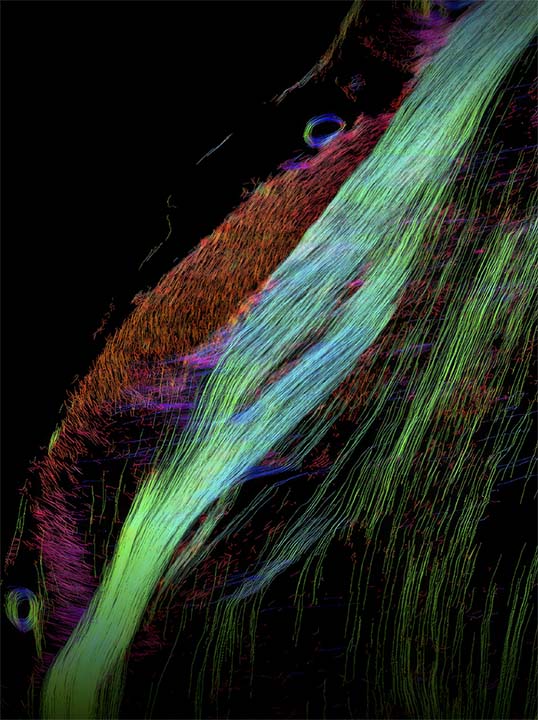
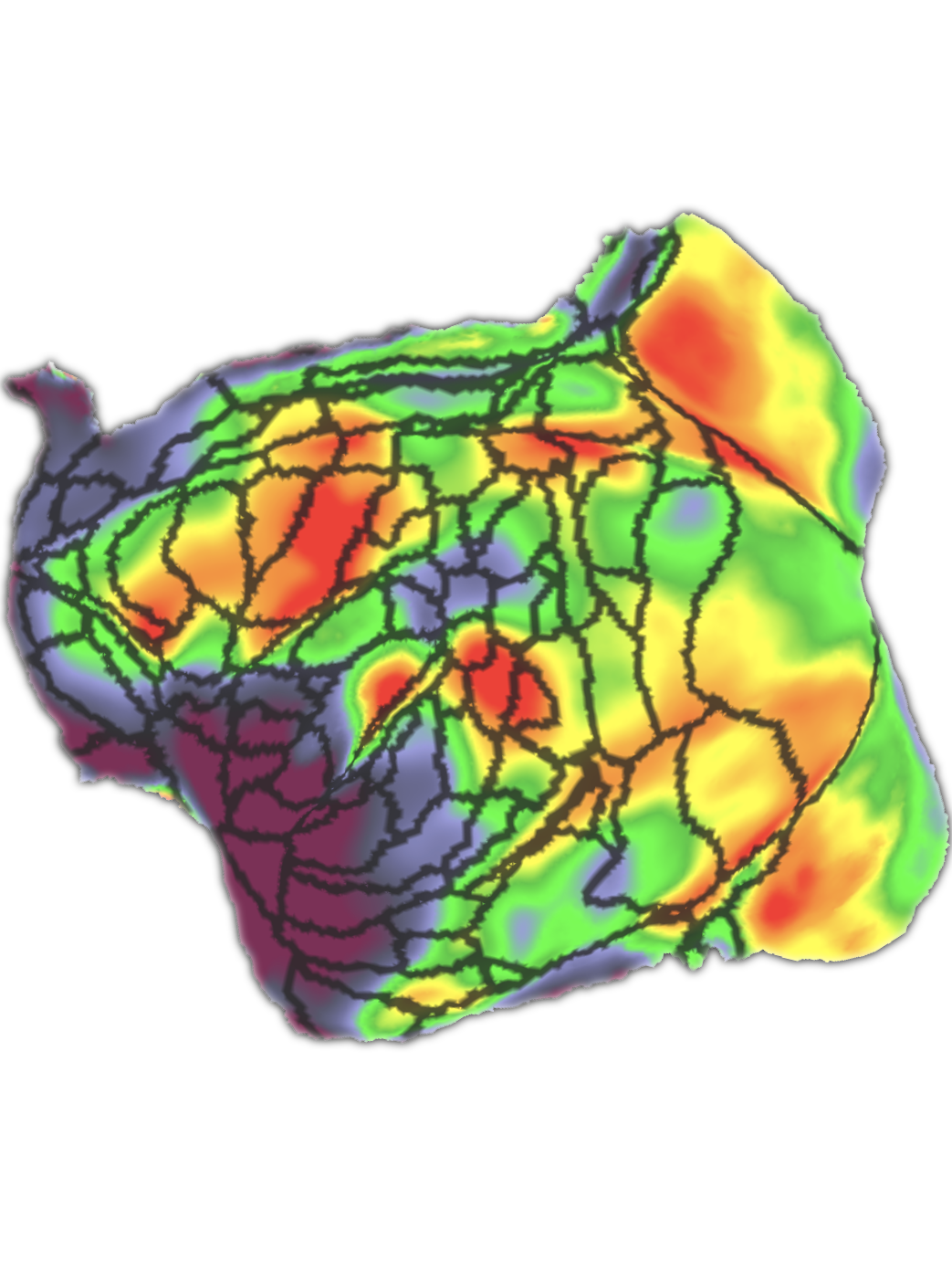
Zhang Y#, Song T#, Yang C#, Shen Y, ..., Poo M, Zhou P, Bi G*, Xiao Y*, Liu C*, Xu F* Whole-brain reconstruction of fiber tracts based on the cytoarchitectonic organization (2025) Nature Methods (*Co-corresponding authors) > Part-2 of the Version-2 (White Matter)@MarmosetBrainMapping.org > A new method (CABLE) to reconstruct whole-brain fiber tracts from cytoarchitecture > Cellular-resolution mapping to disentangle complex intersecting fibers > Validation against brain-wide AAV axon tracing in the same animal > Identification of cell-type contributions to orientation coherence using spatial transcriptomics
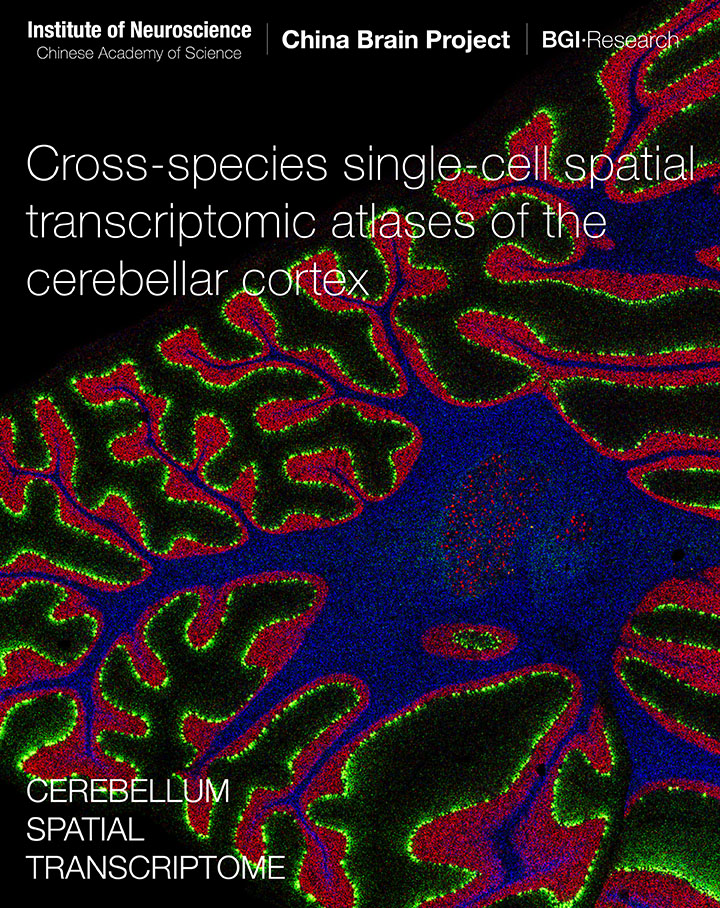
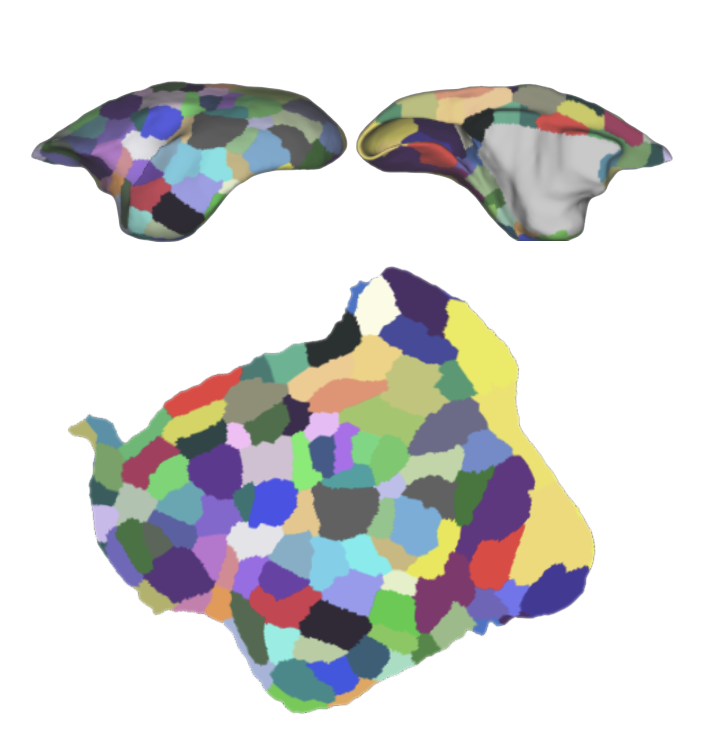
Hao S, Zhu X, Huang Z, Yang Q, Liu H, Wu Y, Zhan Y, Dong Y, Li C, ..., Zeeuw C*, Shen Z*, Liu Z*, Liu L*, Liu S*, Sun Y*, Liu C*. Cross-species single-cell spatial transcriptomic atlases of the cerebellar cortex (2024) Science. doi:10.1126/science.ado3927 (*Co-corresponding authors) > Version-5 series@MarmosetBrainMapping.org > Mapping cell types and gene expression by single-cell spatial transcriptome > Comprehensive cross-species comparsion between primates and mice > Integration of spatial transcriptome and awake fMRI
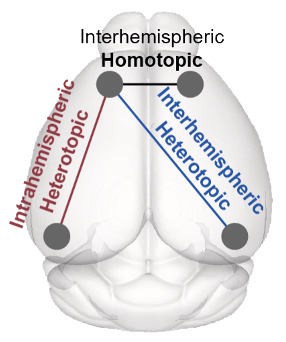
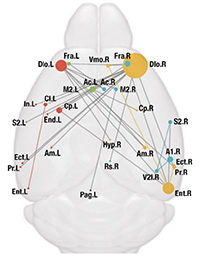
Fei Y, Wu Q, Zhao S*, Song K, Han J*, Liu C*. Diverse and asymmetric patterns of single-neuron projectome in regulating interhemispheric connectivity (2024) Nature Communications doi: 10.1038/s41467-024-47762-y. (*Co-corresponding authors)
> The application and computational modeling of single-neuron projectome > Wide-field calcium imaging of awake and sleep for functional connectivityMeng X, Lin Q, Zeng X, Jiang J, Li M, Luo X, Chen K, Wu H, Yan H, Liu C*, Su B*. Brain developmental and cortical connectivity changes in the transgenic monkeys carrying the human-specific duplicated gene srGAP2C (2023) National Science Review. doi: 10.1093/nsr/nwad281. (*Co-corresponding authors)
> Transgentic monkeys for human brain evolutionZhu X, Yan H, Zhan Y, Feng F, Wei C, Yao Y*, Liu C*. An Anatomical and Connectivity Atlas of the Marmoset Cerebellum (2023) Cell Reports. doi:10.1016/j.celrep.2023.112480 (*Co-corresponding authors) > Version-5 series@MarmosetBrainMapping.org > Cover Paper of Cell Reports (Volume 42, Issue 5)
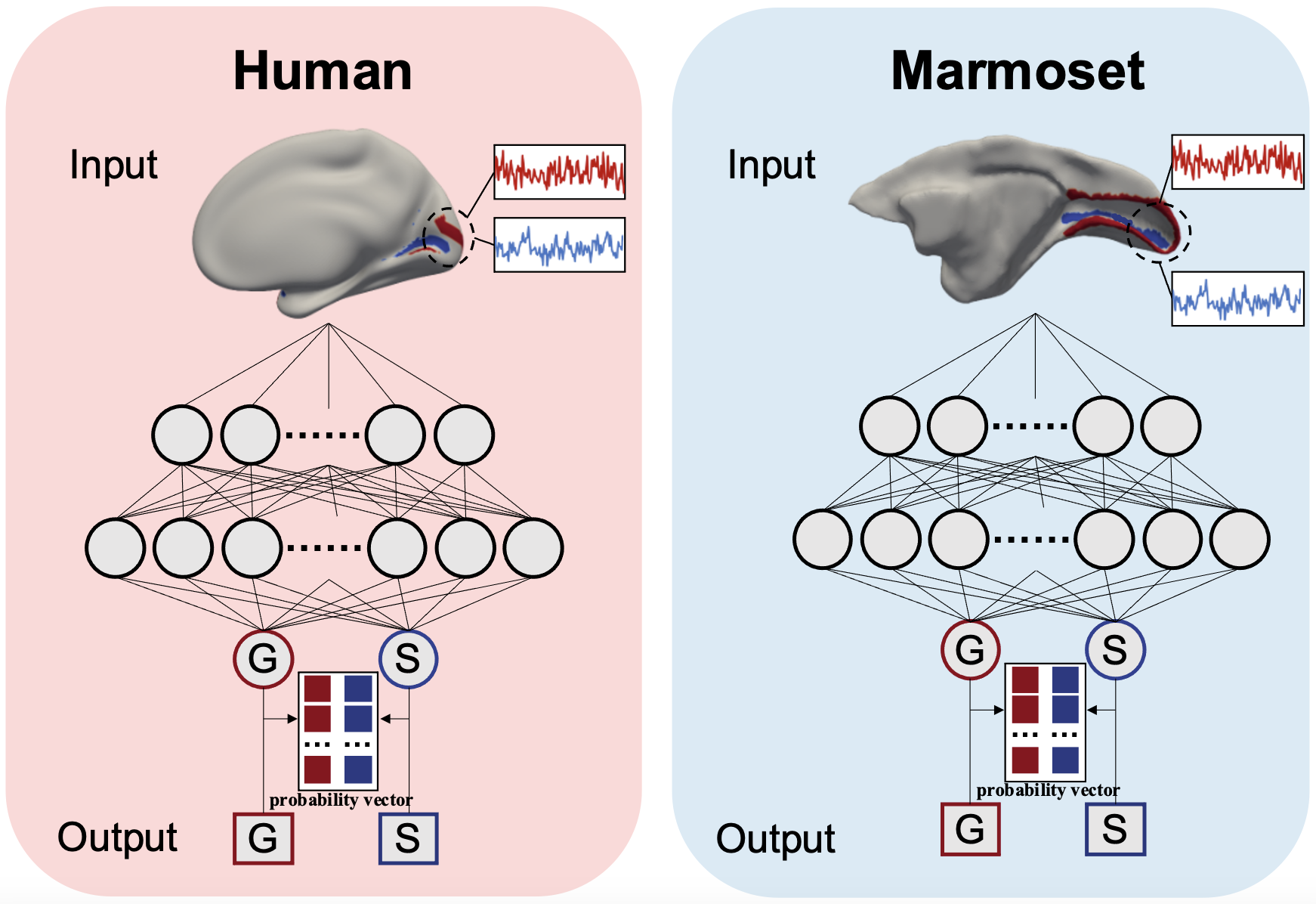
Wang Q, Zhao S*, Liu T, Han J*, Liu C*. Temporal fingerprints of cortical gyrification in marmosets and humans (2023) Cerebral Cortex. doi: 10.1093/cercor/bhad245. (*Co-corresponding authors) > Use Deep Neural Network to identify temporal fingerprints of cortical gyrification
Tian X*, Chen Y, Majka P, Szczupak D, Perl Y, Yen C, Tong C, Feng F, Jiang H, Glen D, Deco G, Rosa M*, Silva A*, Liang Z*, Liu C*. An integrated resource for functional and structural connectivity of the marmoset brain (2022) Nature Communications. doi: 10.1038/s41467-022-35197-2. (*Co-corresponding authors) > Version-4@MarmosetBrainMapping.org > Largest awake resting-state fMRI resource of marmosets
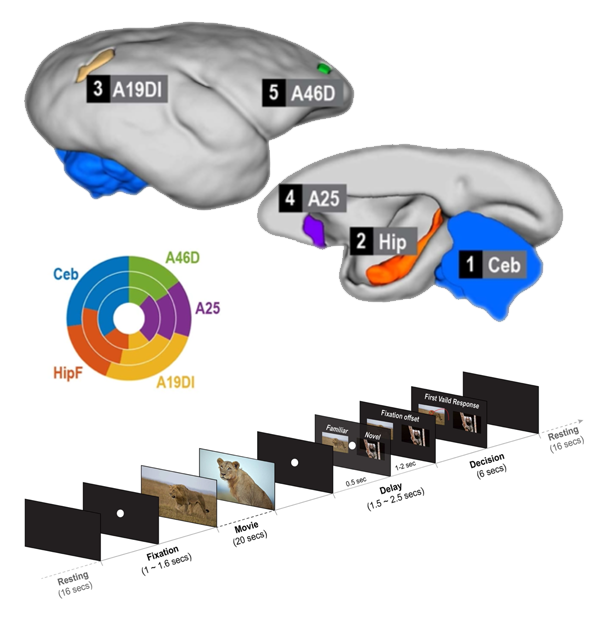
Tian X, Silva A*, Liu C*. The Brain Circuits and Dynamics of Curiosity-Driven Behavior in Naturally Curious Marmosets (2021) Cerebral Cortex. doi: 10.1093/cercor/bhab080. (*Co-corresponding authors) > Task-based (visual) fMRI to explore the innate novelty-seeking brain circuits
Liu C #, Yen C #, Szczupak D, Tian X, Glen D, Silva A*. Marmoset Brain Mapping V3: Population multi-modal standard volumetric and surface-based templates (2021) Neuroimage. doi: 10.1016/j.neuroimage.2020.117620. (#Co-first authors) > Version-3@MarmosetBrainMapping.org > Special Issue (Non-human Primate Neuroimaging) of Neuroimage
Liu C *, Ye F, Newman J, Szczupak D, Tian X, Yen C, Glen D, Majka P, Rosa, M, Leopold D, Silva A*. A resource for the detailed 3D mapping of white matter pathways in the marmoset brain. (2020) Nature Neuroscience. doi:10.1038/s41593-019-0575-0 (*Co-corresponding authors). > Version-2@MarmosetBrainMapping.org > Cover Paper of Nature Neuroscience (Vol23.No.2) > Hightlighted by Nature Methods.
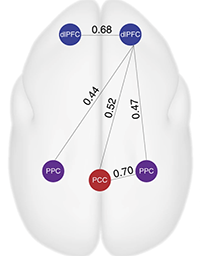
Liu C*, Yen C, Szczupak D, Ye F, Leopold D, Silva A*. Anatomical and functional investigation of the marmoset default mode network .(2019). Nature Communications. doi: 10.1038/s41467-019-09813-7. (*Co-corresponding authors) > Awake functional MRI of Marmoset > Default Mode Network of Marmoset
Shi L# Luo X#, Jiang J#, Chen Y#, Liu C#, Hu T, Li M, Lin Q, Li Y, Huang J, Wang H, Niu Y, Shi Y, Styner M, Wang J, Lu Y, Sun X, Yu H, Ji W*, Su B*. Transgenic rhesus monkeys carrying the human MCPH1 gene copies show human-like neoteny of brain development. (2019). National Science Review. doi: 10.1093/nsr/nwz043. (#Co-first authors)
> Transgentic monkeys for human brain evolutionLiu C, Ye F, Yen C, Newman J, Glen D, Leopold D, Silva A*. A digital 3D atlas of the marmoset brain based on multi-modal MRI. (2018). Neuroimage, doi: 10.1016/j.neuroimage.2017.12.004. > Version-1@MarmosetBrainMapping.org > Cover Paper of Neuroimage (2018 Apr)
Liu C, Li Y, Edwards T, Kurniawan N, Richards L, Jiang T*. Altered structural connectome in adolescent socially isolated mice .(2016). Neuroimage, doi: 10.1016/j.neuroimage.2016.06.037.
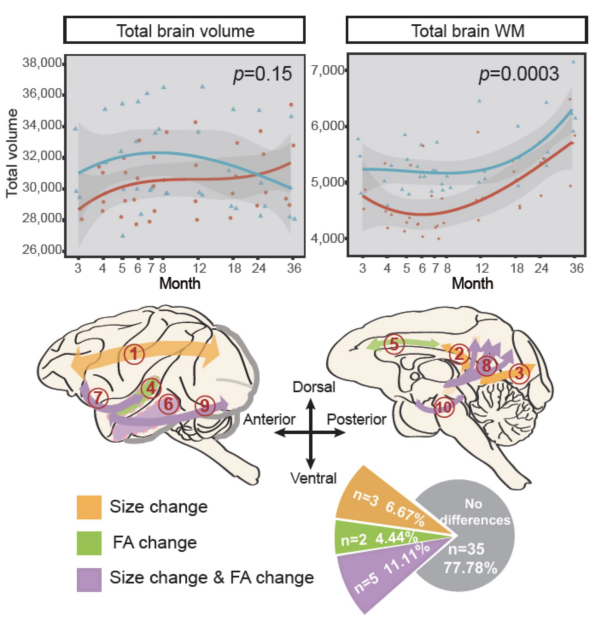
Liu C#, Tian X#, Liu H#, Mo Y, Bai F, Zhao X, Ma Y, Wang J*. Rhesus monkey brain development during late infancy and the effect of phencyclidine: a longitudinal MRI and DTI study. (2015). Neuroimage, doi: 10.1016/j.neuroimage.2014.11.056. (#Co-first authors)